Techniques Used In Genetics
The understanding of the science of genetics has evolved along with the development of several molecular techniques. These techniques are based on basic principles of genetics and are applied to use genetic mechanisms for the benefit of humanity. Overviews of certain techniques are elicited in this chapter. Details of the techniques can be found in standard biotechnology textbooks.
Recombinant DNA Technology
In very basic terms recombination of DNA implies the insertion of fragments or more specifically, insertion of desired genes into certain host cells to utilize the inherent replication mechanisms of the host to produce multiple copies of the gene. This is nothing but cloning of the specific sequences of DNA and thus the process is also termed ‘genetic engineering’.
The specific gene to be cloned may be derived from sources such as another genome of an organism or artificially synthesized in the laboratory. Before the desired fragment of DNA is inserted into a suitable host cell, it is processed by separation from its source. This processed segment has two cut ends that integrate into the host genome and is called the recombinant DNA.
Idea of Recombination from the Nature
Interesting observations on the genetic behavior of bacteria and viruses have inspired the application of those mechanisms to evolve genetic techniques.
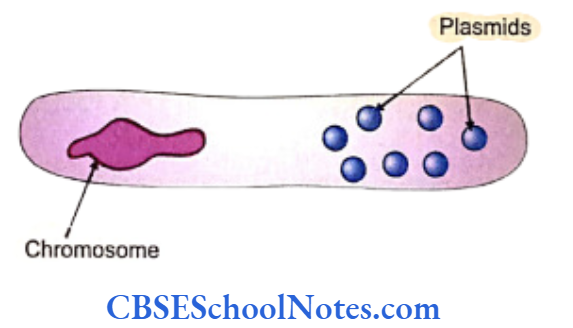
Bacteria and virus are called prokaryotes as they do not have cell nuclei. Eukaryotes are all the other organisms that possess a well-defined nuclear membrane. Bacterial DNA exists in the form of a looped thread-like chromosome or in the form of several smaller ring-shaped genetic material called plasmids. The enormously rapid replication rates of bacteria and virus make them favorites for becoming the host cells in recombination techniques.
Read and Learn More Genetics in Dentistry Notes
Plasmids have the unique property of easily entering a cell and promptly using the cellular mechanism for its replication. Scientists target plasmids for attaching the desired gene to transport them into the host cell. Thus the plasmids act as vectors of DNA.
Though the virus contain nuclear material inside their protein coat, it is mandatory for them to take the help of any other host cell for replication as they lack replicatory enzyme mechanisms of their own. The viruses usually infect bacteria as the host cell, replicate its components inside them, assemble and eventually rupture the bacteria to come out. Recombinant DNA is integrated into viral (bacteriophage) genome and then the virus acts as a vector for the integrated genome.
On the other hand certain enzymes evolve in the bacteria to fight such an invasion by viruses. One of them, the restriction enzymes is used as important tool by scientists in genetic engineering. Viral DNA segments can be fragment at desired sites with the enzyme. This enzyme is used extensively in cleaving required DNA segments from its source. Other enzymes like the ligases are used appropriately to anneal or join ends of DNA fragments.
Process of Recombinant DNA Technique
The sequential procedure of obtaining the desired fragment of DNA (gene) which is to be cloned, multiplication of the obtained gene in a suitable vector, combining the DNA fragment with that of the DNA of vector and transferring of the recombinant vector to the host organism comprise the process of the technique.
Production of the DNA Fragment
The cloning begins by cutting off the DNA at specific sites with enzymes like restriction endonucleases that recognize a set of short DNA sequences (4 to 8 base pair long). The enzymes are named as per their sources, e.g. Eco RI is from the organism E. coli and Hind III from Hemophilus influenzae, and number at about 300.
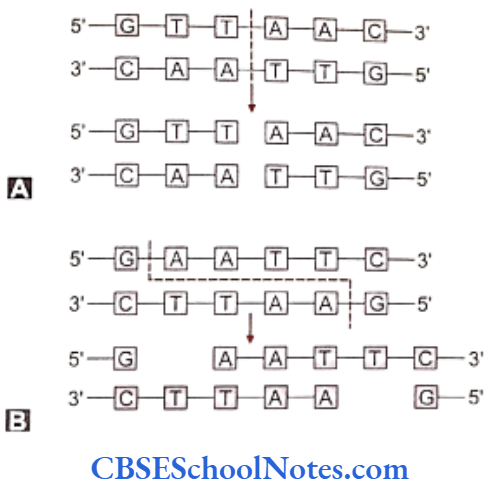
The endonucleases cleave both the strands of DNA but between specific pairs of bases. This cleaving produces either staggered ends or blunt ends at the interface of the cut ends of the DNA. These ends are called ‘sticky’ ends as these cut ends can unite with complementary sequences at any other cut end produced by the same enzyme on a DNA molecule.
Processing of the Vector
Vectors can be obtained from natural sources that can incorporate the desired DNA segment easily into its own genetic environment and transfer the integrated molecule into the host cell for independent and rapid replication. Plasmids, bacteriophages and cosmids are some of the common examples of vectors.
Plasmids, as already mentioned, consist of circular duplex of DNA and occur naturally in bacteria. Plasmids are obtained after disruption of bacteria and then cleaved by restriction enzymes. Restriction enzymes only act at specific sites on the plasmid.
Along with the desired DNA the plasmids also may contain genes expressing factors for antibiotic resistance and thus a particular strain of plasmid can be identified by detecting its resistance against a specific antibiotic.
Integration or Recombination of Desired DNA Fragment with the DNA of Vector
Same restriction enzymes are used to cleave out the desired DNA fragment from its source as well as to break the plasmid between specific bases. This type of breaks in the DNA fragment and in the plasmid’s DNA produces reciprocally complementary ends in both the ‘foreign’ DNA and the cleaved plasmid that combine easily.
As mentioned earlier, the ends of the cleaved DNA are termed ‘sticky’ as they easily combine with cut ends of the plasmid. The enzyme DNA ligase seals and secures the attached ends. The united DNA and the plasmid molecule are then called the recombinant DNA molecule.
Transfer of Recombinant Vector to Host Organism
After the recombination of the DNA fragment and the plasmid, this particle is introduced into the host cell by increasing the porosity of the cell membrane with the application of certain chemicals or high electric voltage across its membrane.
The recombinant molecule starts replication within the cell along with the nuclear material of the cell at each cycle of cell division. Eventually hundreds and thousands of copies of the desired DNA are produced with the help of the host cell machinery.
Screening of Recombinant Vectors
The host cell or bacteria does not accept all plasmids. This selectivity of acceptance creates two kinds of bacteria in the culture media-one type containing the DNA fragment whereas the other not containing it.
The plasmids contain certain genes along their genome that impart them resistance against certain antibiotics. In case the foreign DNA is inserted into the plasmid by cleaving a particular gene responsible for developing resistance for a particular antibiotic, such resistance would be lost in this plasmid though the same plasmid would maintain resistance against some other antibiotics, the genes for which remain intact.
This property of loss of resistance due to recombination is utilized for the detection of bacteria that have accepted the DNA fragments. Separate colonies of bacteria are exposed to the recombination. Bacteria from these colonies are cultured separately and representative bacteria from individual colonies are tested for susceptibility for different antibiotic.
The colony in the subculture showing susceptibility to a particular antibiotic specifies the cleavage in the plasmid and integration of the DNA fragment. These colonies from the master (primary) plate are picked and cultured separately and will contain only bacteria (host cells) with recombinant vectors (plasmids).
Screening of Clones with Specific DNA Sequence
The detection of the recombinant DNA integrated bacterium can be pursued with more refined techniques including nucleic acid hybridization. This method entails direct hybridization of labeled probes on to specific sequences on the recombinant molecule. The identified bacteria are isolated and culture to obtain the desired DNA segment.
The recombinant DNA molecules are thus generated and collected and this collection constitutes the DNA library.
Some Important Applications of Recombinant DNA Technology
Application of the recombinant DNA technology is widely accepted now as an important tool for several useful purposes such as:
- Preparation of chromosome maps and analysis of DNA sequences.
- Production of drugs like insulin, somatostatin, blood clotting factors, growth hormones, synthetic vaccines like antirabies, antimalarial, antihepatitis and cholera vaccines, interferon from genetically engineered E. coli to combat viral infections and monoclonal antibodies against certain organisms.
- Using in the diagnosis of genetic diseases and gene therapy.
Polymerase Chain Reaction
The amplification of DNA sequences (genes) described in the above sections is based on utilization of the host cellular mechanisms and thus is known as “in vivo” cell-based cloning. Genes can be cloned by non cellular “in vitro” techniques such as the polymerase chain reaction (PCR) that is done in machines.
Copies of DNA sequences can be produced in large amounts with PCR. The essential prerequisite for this technique is that we must know the sequences of the DNA of the either sides (flanking regions) of the desired segment to be cloned. This knowledge is mandatory for the formulation of the ‘primers’ (discussed later). Only a very small amount of DNA (even of a single cell) is needed to produce millions of copies of the DNA fragment.
Concept of PCR Technique
As shown in the figure below, DNA replication needs at least a single molecule of a double stranded DNA to begin with. The two strands are separated (denatured) with regulation of temperature. Enzymes, nucleotides and primers are then added to make-up a mixture in the PCR machine (thermo-cyclers). Once added, the nucleotides get arranged on each of the denatured DNA single strand. Thus the new complementary strand along with the old strand together forms the double helix.
The nucleo-tides are attached one by one to the primer CLEIC ACID PROBES to their 3’ends. The primers, as seen in the figure below, are attached one on each of the denatured starting molecule of the DNA at opposite ends. In the PCR technique, the primers (short nucleotide stretches) are added in the machine along with enzymes and the nucleotides. The primers are actually very short DNA sequences called deoxyoligo-nucleotides.
DNA polymerase as well as the four nucleotides are added to the cloning mixture. It is therefore essential to have an idea of the flanking sequences of the desired gene to be cloned in order to produce the primers. These primers essentially limit the stretch of the big DNA molecule to be replicated. The DNA molecule confined between the primers is acted upon by the artificial cloning machinery to replicate the trapped segment of the DNA.
The DNA fragments, the primers, the oligonucle-otides and DNA polymerase enzyme (the heat stable “Taq polymerase” derived from the Thermus aquaticus) are all incubated in the machine and the required temperature for amplification is regulated externally.
Each cycle of replication is repeated with fresh denaturation of the double helix and annealing of added nucleotides to the annealed primers. This results in the replication of a DNA segment in an exponential proportion. PCR thermal cyclers are automatic and need not be set again after each round of amplification. Modification of techniques can also produce mutations in DNA fragment, as desired.
PCR is a valuable technique used for detecting infectious agents like viruses, for prenatal investigations, tissue typing for transplantation, studying polymorphisms, evolution and several other applications.
Nucleic Acid Probes
Radiolabeled probes help to recognize complementary sequences in DNA or RNA molecule. This helps to identify and isolate the specific DNA sequences from an organism. Nucleic acid probes are small stretches of DNA that can be derived from various sources. These probes anneal to complementary sequences, if these complementary target sequences are present in the sample DNA.


These probes also help in the diagnosis of infectious diseases and identification of specific causal organisms. Forensic tests (DNA fingerprinting) are based on the same principle.
Detection of DNA Segments with Nucleic Acid Hybridization
The following steps are followed sequentially to identify DNA segments of interest from a given genomic population
- After the DNA molecules are extracted, they are digested with application of restriction enzyme so that they are cleaved into multiple segments of different sizes.
- The DNA sample is run in electrophoresis where the fragments are arranged according to their sizes along the gel.
- Bands appear on the gel at specific intervals depending on the molecular weights of the fragments.
- These bands are stained and visualized directly in the gel.
- These bands can be isolated for analyzing their DNA sequences. A particular gene (DNA segment) can be identified within those bands with the help of radio tagged molecular probes that bind to definite denatured strand.
A particular segment in a band in the gel can be identified by hybridization with molecular probes. This process requires transferring of the band from the gel to a nitrocellulose paper. This transferring technique is called ‘blotting’. Several types of blotting are enumerated according to the involvement of different molecules.
Blotting of DNA bands on nitrocellulose paper- Southern blotting.
Blotting of mRNA bands on nitrocellulose- Northern blotting.
Blotting of protein on nitrocellulose membrane- Western blotting.
Southern Blotting
Southern blotting is one of the most used techniques in genetics. DNA is extracted from a source and then digested using specific restriction enzymes that cut the DNA strand into several smaller segments at sites specific to the enzymes. This sample is then run in a gel electrophoresis equipment. The bands thus formed on the gel are then denatured with alkali.
This gel is now placed between a buffer saturated paper and a sheet of nitrocellulose membrane. The movement of the buffer from the paper to the nitrocellulose membrane passes through the gel and carries the denatured single strands of DNA from the gel to the membrane (blotting). The transferred DNA is now fixed on the membrane by heating it at 80°C for 2 to 3 hours.
The membrane can now be subjected to exposure to radio-labeled probes that hybridize to specific sequences in the DNA. After proper washing of the membrane, an autoradiograph of hybridized DNA may be taken on an X-ray film for the presence of the desired strand of molecule in any given band on the membrane.
The appearance of a band at a particular level will happen only when a DNA segment of the particular length is present in the sample after the application of restriction enzyme. Similarly, attachment of specific probes will depend upon the presence of the particular DNA segment complementary to the probe.
Bands at a particular level denote the presence of similar fragments that can be compared with a given reference. Bands may be of different thickness at the same levels of reference and across the length of the gel.
DNA Sequence Of Gene Or A DNA Segment
As an extension to the step of DNA isolation and segment identification, one may proceed to determine the sequencing of nucleotides on the DNA molecule (DNA sequencing). Several methods are utilized as tools for sequencing and are designed on the basis of different principles of genetics.
The most commonly used technique has been the dideoxy chain termination method. The more recent automatic sequencers apply a variant of this method.
Dideoxy Chain Termination Method (enzymatic)
This is one of the earliest processes employed in the late 70’s to determine DNA sequences. Denatured single stranded DNA fragments that are to be sequenced are taken in four different reaction tubes as the first step. All the tubes contain several identical copies of fragments of DNA molecules to be sequenced.
Radioactively labeled four different deoxy- nucleotides, enzyme DNA polymerase I and oligonucleotide primers all are added to each of the four tubes. The deoxynucleotides are molecules that anneal against complementary nucleotides on the denatured DNA strand and maintain the elongation of the new strand that is being synthesized.
Each tube also receives one of the four dideoxynucleotides and as such each tube has a different dideoxynucleotide. The dideoxynucleotide molecules are different from the deoxynucleotide molecules as the former lack in a hydroxyl group in one of their carbon atoms. The attachment of a dideoxynucleotide to the growing strand immediately stops the chain elongation.
This termination of chain elongation is randomly affected. This means that the termination of chain elongation depends on the ‘chance’ of attachment of the particular type of a dideoxynucleotide to its complementary nucleotide in the template strand. Therefore, in a given tube we can find millions of chains of different lengths terminated randomly on the event of attachment with the specific dideoxynucleotide in the tube .
Once (in a given tube) the particular dideoxy- nucleotides are incorporated in the chain with stopping of chain elongation, the fragments are taken out from the tube and run in electrophoresis. Terminated chains in all the four tubes are run in electrophoresis equipment on four adjoining lanes, each lane denoting chain terminations due to attachment of four different dideoxynucleotides. These attachments are random, giving rise to chains of different lengths that arrange according to their lengths in the gel.
From this information we can identify the particular terminal dideoxynucleotide that stops chain elongation and hence identify the complementary terminal nucleotide in that chain. In this way all the terminal nucleotides in all the chain fragments in all the four lanes can be computed in a sequence yielding precise sequencing of a DNA molecule. Autoradiographic methods are applied in detection of the radiolabeled dideoxynucleotides in the electrophoretic bands.
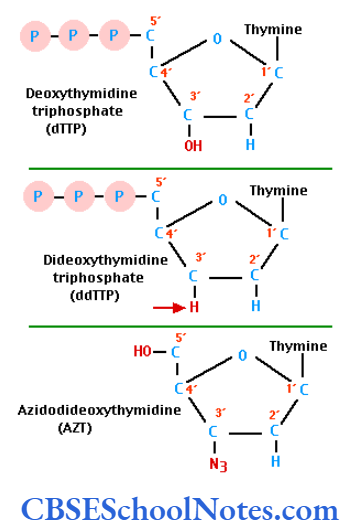
Automatic Sequencers
Modern day automatic sequencing machines are computerized and highly accurate and rapid. Different fluorescent dyes are attached to the oligonucleotide primer in each of the four reaction tubes. The resultant gel mixture is then electrophoresed in a single gel tube instead of four.
A fluorescence detector measures the color from the gel tube and automatically records sequences. This method is of course a further modification of the dideoxy process. The credit for rapidity with which the human genome project was conducted goes to these automatic sequencers.
DNA Fingerprinting
In all humans the genome comprises of the coding as well as noncoding regions. In the noncoding DNA regions the sequences are very repetitive and are called tandemly repeated DNA sequences. The collection of these repetitive sequences imparts unique identities to individuals. The pattern of occurrence, length and number of these repeats are unique and specific for each individual. The concept of DNA fingerprinting is based on the above principle.
DNA fingerprint is an important tool for identification of individuals, settlement of disputed paternity, criminal investigations and related purposes. DNA fingerprints in identical (mono- zygotic) twins are exactly the same.
The DNA Fingerprinting Technique
The technique begins with obtaining DNA from a source that may be as varied as the body fluids, cells or sequestrated dead tissues often several years old (DNA is usually a very stable molecule). Obtained DNA is cleaved into smaller fragments with the help of endonuclease enzymes.
The action of the endonuclease enzyme differs in individuals as the enzyme cuts individual genomes at different places due to the presence different patterns of the tandem repeats sequences in different individuals. These ununiform cuts in the genome give rise to DNA fragments of different lengths in individuals. The fragments of DNA are subsequently separated by agarose gel electrophoresis. Southern blotting is then applied to transfer the bands on to nitrocellulose.
As mentioned earlier, variability in individual genomes occurs specially in the noncoding regions in the forms of several polymorphisms. These polymorphisms exist as short (2-3 bp), inherited tandem sequence repeats like CACACA……that run repeatedly at several locations (loci) in the genome. The sequences are called microsatellites which occur throughout the genome at microsatellite loci. The numbers of such tandem sequences running at each locus may vary from 4 to 40 in different individuals.
This variation in the number of nucleotide runs at these loci is called Variable Number of Tandem Repeats (VNTR) or a hypervariable satellite sequence. The numbers of these variations are different in individuals and when several of these loci are taken into account, the diversity between individuals is enormous and the chance of two random individuals sharing the same genetic pattern is one in a billion.
Primers that bracket important loci with VNTRS have been developed. These bracketed variable sequences can be amplified with the help of PCR. A set of 5 to 10 such VNTR loci are chosen and amplified for the complete comparison between individuals. The band patterns of these variable sequences produced after electrophoresis are akin to the molecular fingerprint of an individual.
As discussed earlier, the presence of these variable sequences also alter the sites of action of restriction enzymes. Action of these enzymes on different individual genomes would cut the DNA at different places in different individuals. This polymorphism is called the Restriction Fragment Length Polymorphism (RFLP).
RFLP in the DNA produce bands at different levels (according to the different lengths of DNA segments) in the electrophoretic gel. This property of variance is used to detect the molecular finger- printing of a person or used to detect associations of a type of RLFP with a disease.
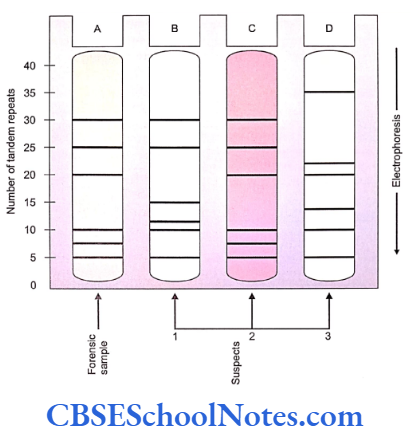
In the given example, a sample from the site of crime (e.g. a strand of hair from the victim’s nails) is amplified by PCR. A set of 3 variable loci are selected and amplified from the sample. Since each of the loci is represented in two homologous chromosomes, 6 bands are obtained after the amplification. Samples are collected from the suspects and the same loci are amplified and banded.
Identification of the culprit (individual 2 in the example) is accurate using this technique when the overall pattern of its bands matches significantly with that with the sample. This approach can be used in the testing of paternity. In cases of paternity disputes matching of at least half of the bands usually settles the issue.
Human Genome Project
A monumental project was started in 1991 in the USA involving 16 laboratories, 1,100 biologists, computer scientists and technicians from countries like USA, UK, France, Japan and others. The design behind the human genome project (HGP) was to sequence the entire human genome with a view to provideing an extensive understanding of DNA sequences, the organization, function and evolution of the human genome, to map both normal and anomalous disease- specific genes, give boost to functional and comparative genomics and bioinformatics.
Constant upgradation of techniques helped to complete this public funded project in February 2001; well before the stipulated time of 15 years. Two research facilities the Celera Genomic Corporation and the HGP working on the project released their data simultaneously to the world.
The project yielded several important observations about the human genome. About 3.2 billion base pairs constitute the human genome with approximately 30,000 genes in contrast to the earlier concept of presence of greater number of genes in the genome. This finding, supplemented with further studies in proteomics, has forced us to reconsider the validity of the “one gene-one function (enzyme)” concept in favor of a better explanation for protein synthesis. The genes comprise only about 5% of the genomic bulk. Rest of the genome is made up of noncoding DNA (junk DNA) containing segments of repetitive DNA.
Human DNA can be dated (DNA dating) with the analysis of such repeats and family trees can be assembled that explain the source, the point of first occurrence and subsequent evolution and dispersion of any segment of the genome in an individual, farly, clan or an entire race.
It is interesting to observe that several bacterial genes have been introduced by nature directly into the human genome without undergoing the grind of evolution.
Though smeared with its share of controversies, the HGP was a historical and colossal endeavor in Though smeared with its share of controversies, science that has brought forth vast data and understanding that would eventually benefit the mankind, with its proper utilization.
Stem Cell Research
Stem cells are those cells that are capable of self- renewal (can divide to produce cells with same properties) and also are able to differentiate into specific types (lineages) of cells. Thus stem cells are basically pluripotent and multipotent (if not totipotent) progenitor cells that not only divide to produce cells of the same nature but given the appropriate environment, they differentiate into other tissues as well.
It is evident that potency of cells diminishes as an individual grows from a zygote towards an adult. Therefore, it is understood that early embryos are the best sources of stem cells (embryonic stem cells). Stem cells can also be harvested from the umbilical cord at a later stage of life (cord blood stem cells).
In adults the bone marrow is a good source of stem cells (mesenchymal stem cells). It is to be noted that the potency of the stem cells depends on its source. After collection of the stem cell rich tissue, the stem cells are separated by various cell sorting techniques using sophisticated equipment as per the requirements. The stem cells bear several cell markers on their cell walls that give them specific identities.
Cells are preserved at very low temperatures (cryopreservation). When desired the cells can be thawed back to normal conditions. Cells can be then cultured for multiplication or put in an environment that leads the cells to differentiate into required progenitor cells. These cells are then injected into damaged tissues where they are thought to replicate and repair the damaged organs.
Embryonic stem cells are good models to study the effects of bacterial toxins, drugs, etc. One can preserve his/her own cord blood stem cells (banking) in case of a need to him (autologous transplantation) or others (allogenic transplantation) in the future. An important aspect of stem cell therapy is that it does not need stringent compatibility match for allogenic transplantations.
Prenatal Diagnosis, Techniques And Genetic Counselling
The term “Prenatal diagnosis” is used to define the process of detecting a disease or the risk of occurrence of a disease in humans. These tests are done well before the birth of the fetus. This approach of detecting severe debilitating or fatal diseases proves to be very helpful in pregnancies with a high risk for any such genetic disease. This gives an opportunity to the couple to decide the fate of the affected pregnancy before it reaches to a very advanced stage.
Prenatal detection of dental diseases is not practiced. Prenatal diagnosis of congenital and other genetic disorders is common in cases of diseases that are incompatible to life or diseases that cause severe physical and mental disability. Detection of dental disorders may be a matter of correlation or coincidence occurring as a part of these systemic diseases.
Prenatal detection of diseases is advised in cases of high-risk pregnancies with the history of previous incidence of the disease in the family.
Indications for Prenatal Diagnosis and Techniques
Couples having a history of any genetic disorder or undiagnosed physical abnormality in the family are ideal candidates for prenatal diagnosis of the fetus. History of a neural tube defect in the family or any genetic syndrome in a previous child, pregnant women of 35 years or above calls for a prenatal diagnostic test in the unborn child. In case of suspicion of an X-linked recessive disorder, mothers should be screened for their carrier status.
Few of the prenatal diagnostic techniques are summarized below:
Amniocentesis
Amniocentesis entails the process of collection of fetal cells for chromosomal analysis. The technique is preferably applied between 14-16 weeks of gestation. 10 to 20 ml of amniotic fluid is tapped through the abdominal wall of the mother and the cells that are collected are used for karyotyping (Refer Chapter 3).
Neural tube defects can be diagnosed in the prenatal state by estimation of a-fetoprotein levels in the amniotic fluid. Its level is raised in the amniotic fluid as a result of leakage from the open neural tube defects. The procedure of amniocentesis carries only 1% risk of abortion.
Chorionic Villus Sampling
Chorionic villi samples are aspirated with a catheter introduced through the cervix under strict asepsis and ultrasound guidance. Analysis of the cells is possible even without culturing them as the cells in these tissues grow rapidly. The biopsy of the tissue is used for biochemical assay or DNA analysis to detect any genetic disorder.
Chorionic villus sampling is preferably done during 10 to 11 weeks of pregnancy, i.e. a few weeks earlier than the designated period for an amniocentesis (14-16 weeks). The risk of abortion is enhanced due to the small gestation size and manipulation of the vital villus structures. It is rated at 2 to 3% and is higher than that with amniocentesis. The procedure may be associated with limb anomalies if carried out prior to 9 weeks of gestation.
Ultrasonography
It is a safe method of prenatal diagnosis both for the fetus and the mother. It is routinely performed at 12 weeks of pregnancy for detection of multiple pregnancies, fetal malformations, fetal age determination and classification of the type of placenta. Nuchal translucency (NT), exomphalos, rocker- bottom foot, etc. are some of the features related to certain chromosomal abnormalities detected with ultrasonography.
Serum Screening in the Mother and Blood Sampling in the Fetus
Detection of certain elements in maternal blood sample may act as clues for a probable disease in the offspring. The presence of “alpha-feto-protein” (AFP) in maternal serum at 16 weeks gestation indicates towards a neural tube defect (anencephaly or spina bifida).
Fetal blood is drawn under ultrasound guidance from one of the umbilical vessels by putting a transabdominal percutaneous needle into the mother’s abdomen. The procedure is often referred to as “cordocentesis”. The indication of this procedure is to use the collected fetal blood to arrive at a prenatal diagnosis of blood disorders and for chromosomal analysis. A high risk of abortion (about 10%) is associated with the procedure.
Genetic Counselling
At the existing levels of medical understanding and therapeutics the scope of curing genetically incurred diseases is quite remote. Interventions in terms of replacement of defective genes or their products by genetic engineering are not commonly practiced. Therefore, the knowledge of the principles of genetics can be used to detect the risk of incidence of some of the genetic diseases. This information, then, can be used to prevent their occurrences or reduce the severity of their outcome.
The services of a counselor should be sought by patients distressed with genetic disorders to help them attenuate their sufferings or couples, who are at risk of having a genetically abnormal child, to consider specific detection and treatment measures. The counselor should provide realistic and. appropriate suggestions about the disease.
Diagnosis Of Genetic Disease
It is vital that the patient and his relatives should be informed about the correct diagnosis of the disease as well as the mode of its inheritance. The risks for occurrence of the disease are calculated on the basis of the understanding of laws of Mendelian inheritance. The prognosis and availability of treatment, if any, should also be positively discussed. Arriving at a diagnosis may involve 3 pertinent steps.
- Family history
One should make a detailed pedigree chart to analyze the mode of inheritance of the trait or disease. - Examination of patient
A careful clinical examination of patient will help to reach a correct diagnosis. - Laboratory investigations
This may include biochemical investigations, chromosomal analysis and molecular studies. Antenatal diagnostic interventions may be sought in high-risk pregnancies, e.g. amniocentesis, chorionic villi sampling, imaging, etc. that would aid correct diagnosis.
Management Of Genetic Disease
Parents as well as individuals should be clearly told about the diagnosis and the risk of occurrence or recurrence (as applicable) involved in case they decide to continue an affected pregnancy or to have children in future. A counselor should also tell them about all the options available for the management of any genetic disease.
As most of the genetic disorders are incurable, one should try to prevent or limit the disabilities of the disorder. Couples left with no other alternatives of having a normal child, should be recommend to think for an adoption. Subsequent to a positive prenatal detection and all explanations given by the counselor, the decision regarding the fate of the pregnancy has to be made only by the couple involved.
